TL;DR:
- CLAMP introduces a novel AI architecture for activity prediction in drug discovery.
- It utilizes contrastive language-assay-molecule pre-training to improve predictive performance.
- The model offers zero-shot transfer learning capabilities, providing insightful predictions for new bioassays.
- CLAMP’s modular architecture and pre-training objective contribute to its remarkable success.
- The tool revolutionizes molecular activity prediction, bringing exciting possibilities to the drug discovery industry.
Main AI News:
In the vast realm of scientific research, one enduring challenge has been accurately predicting a molecule’s chemical, macroscopic, or biological properties based on its chemical structure. Over the years, significant technological advancements have led to the application of various machine learning algorithms to discover correlations between the chemical makeup of molecules and their characteristics. Notably, the emergence of deep learning brought about activity prediction models, which play a crucial role in the computational drug discovery industry, akin to large language models in natural language processing and image classification models in computer vision. These deep learning-based activity prediction models utilize diverse low-level chemical structure descriptions, such as chemical fingerprints, descriptors, molecular graphs, SMILES representations, or combinations thereof.
While these architectures have shown promise, their progress has not been as revolutionary as their counterparts in vision and language domains. Typically, training these activity prediction models involves pairs of molecules and activity labels from biological experiments, known as “bioassays.” However, annotating such training data (bioactivities) is extremely time-consuming and labor-intensive. As a result, researchers are actively seeking methods that can efficiently train activity prediction models with a reduced number of data points. Furthermore, current activity prediction algorithms face limitations in effectively utilizing comprehensive information about the prediction tasks, primarily presented in the form of textual descriptions of biological experiments. The models’ reliance on measurement data from the bioassay or activity prediction task they are trained on hampers their ability to perform zero-shot activity prediction and yields poor predictive accuracy in few-shot scenarios.
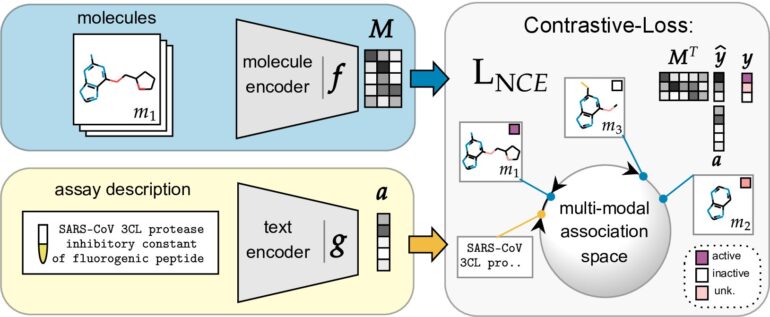
Addressing these challenges, a distinguished group of researchers from the Machine Learning Department at Johannes Kepler University Linz, Austria, have proposed an innovative solution: the Contrastive Language-Assay-Molecule Pre-training (CLAMP) architecture for activity prediction. CLAMP offers the unprecedented capability to condition its predictions on textual descriptions of the prediction task. This groundbreaking architecture comprises distinct molecule and language encoders, pre-trained in a contrastive manner across these two data modalities. To improve activity prediction, the researchers recommend using chemical databases as training or pre-training data and adopting an efficient molecule encoder. Such databases offer an abundance of chemical structures, far surpassing those found in biomedical texts.
At the core of CLAMP’s operation is a trainable text encoder responsible for generating bioassay embeddings and a trainable molecule encoder for creating molecule embeddings. These embeddings are normalized at the layer level. The Austrian researchers have also incorporated a scoring function that yields high values when a molecule exhibits activity on a specific bioassay and low values otherwise. Additionally, the contrastive learning strategy equips the model with zero-shot transfer learning capabilities, enabling insightful predictions for unseen bioassays. Extensive experimental evaluations conducted by the researchers showcased significant enhancements in predictive performance on few-shot learning benchmarks and zero-shot problems in drug discovery, along with the generation of transferable representations. The model’s remarkable success is attributed to its modular architecture and pre-training objective.
While CLAMP represents a major leap forward in activity prediction, it is important to acknowledge areas for potential improvement. Factors like chemical dosage, which may influence bioassay results, are currently not considered. Additionally, there may be instances of incorrect predictions stemming from grammatical inconsistencies and negations. Nevertheless, the contrastive learning approach of CLAMP demonstrates superior performance in zero-shot prediction for drug discovery tasks across various extensive datasets.
Conclusion:
The introduction of CLAMP, a groundbreaking AI tool for molecular activity prediction, marks a significant advancement in the drug discovery market. Its ability to adapt to new experiments, excel in zero-shot learning scenarios, and enhance predictive performance opens up new avenues for efficient drug development processes. With CLAMP’s promising results and potential for further improvement, pharmaceutical companies and researchers can now approach drug discovery with greater confidence and effectiveness, accelerating innovation and driving growth in the market.