TL;DR:
- CT-SLEB, a novel machine learning method, enhances genetic risk assessment for non-white populations.
- Existing risk scores are less accurate for minority populations.
- Researchers from Johns Hopkins, Harvard, and the National Cancer Institute developed CT-SLEB, combining multiple techniques.
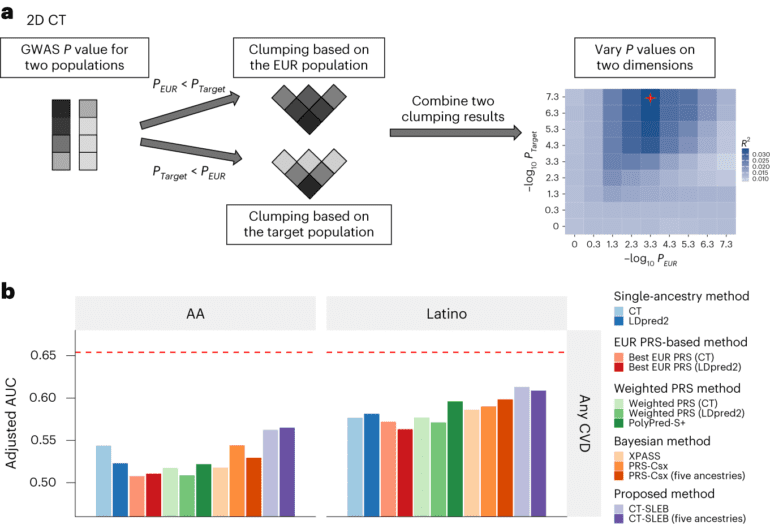
- CT-SLEB analyzed 19 million genetic variants from 5 million individuals, including minorities.
- Promising results suggest improved accuracy and scalability for diverse populations.
- Genetic risk scores can help target interventions, but disparities exist.
- Addressing biases in algorithms and data representation is crucial.
- CT-SLEB offers quick runtime and scalability for larger datasets.
- The software code is publicly available.
- Collaboration with 23andMe provided unique access to diverse datasets.
Main AI News:
In the realm of medical science, the role of genetics in the development of various diseases and chronic conditions is undeniable. While certain genetic disorders, like cystic fibrosis or sickle cell anemia, stem from single-gene variations, complex or polygenic diseases such as cancer, type 2 diabetes, and heart disease result from an intricate interplay of multiple gene variations alongside environmental factors.
Polygenic risk scores, abbreviated as PRS, serve as indicators of an individual’s genetic susceptibility to a specific disease – higher PRS values signify a heightened likelihood of developing that disease. A critical limitation, however, has been the reliance on data primarily from individuals of European ancestry to develop and evaluate existing PRS models. As a consequence, these risk scores and the algorithms used to compute them lack precision when applied to underrepresented minority populations, particularly those of African descent.
In response to this challenge, a groundbreaking solution has emerged through collaborative efforts between researchers at Johns Hopkins University, the Harvard School of Public Health, and the National Cancer Institute. Their innovation, known as CT-SLEB, is meticulously detailed in a paper featured in Nature Genetics.
Nilanjan Chatterjee, the Bloomberg Distinguished Professor of biostatistics and genetic epidemiology at Johns Hopkins, oversaw this pioneering study. He underscores the importance of not only advancing computational methods but also collecting extensive data from diverse populations. Chatterjee states, “Better methods are important, but those alone will not eliminate the performance gap of these models across diverse populations. We also need to collect more data on various minority populations so that these models can be better trained and the accuracy of genetic scores for these populations is improved.”
The CT-SLEB method amalgamates elements from several existing techniques, including the clumping and thresholding (CT) method, an empirical-Bayes (EB) approach, and a super-learning (SL) model. In their analysis, the researchers scrutinized over 19 million genetic variants from data pertaining to more than 5 million individuals, including a substantial representation from underresearched minority populations. In large-scale genome-wide association studies across diverse ancestral backgrounds, CT-SLEB exhibited promising performance and scalability, indicating its potential for future implementation.
Genetic risk scores hold immense promise in identifying individuals who may benefit from targeted interventions due to their heightened risk for specific diseases. Nevertheless, disparities in the performance of polygenic risk scores across different populations have raised concerns about their clinical application potentially exacerbating healthcare inequalities.
Chatterjee underscores this point, saying, “The problem is the genetic scores that have been derived so far don’t perform as well in African ancestry populations. As we know, there are already a lot of barriers that lead to healthcare disparities, and we don’t want to introduce another. We need to push toward collecting more data and developing better algorithms to allow for an equitable approach to bringing this new technology into the field in a way that is inclusive and benefits everybody.”
The study’s lead author, Haoyu Zhang, emphasizes the broad implications of their findings for enhancing health equity through improved polygenic risk predictions across diverse populations. Zhang notes, “Better PRS performance can lead to more accurate risk prediction, which facilitates early disease detection, prevention, and personalized treatment strategies.”
Chatterjee also underscores the importance of addressing racial biases in algorithms, including those developed through artificial intelligence methods. He emphasizes the need to identify and rectify these biases early on to prevent further societal disparities. He says, “With AI and machine learning algorithms, you have to be really careful about these biases being introduced early on because once they become deeply entrenched into the system, it is difficult to change practice.”
Elizabeth Stuart, chair of the Department of Biostatistics at the Bloomberg School, further highlights the significance of considering data sources and representation. She stresses, “This work highlights the importance of paying attention to where data comes from, who is represented, and, crucially, who is not represented, in traditional data sources.”
In addition to its enhanced performance for diverse populations, CT-SLEB boasts a swift runtime, enabling it to analyze data and compute polygenic risk scores at a faster pace than other methods. It demonstrates the potential for scalability to handle larger datasets in the future, accommodating more genetic variants and diverse populations. The software code for CT-SLEB and all related analyses are publicly available on GitHub. The team is actively working on additional methods that are computationally efficient and can further enhance risk score accuracy through advanced modeling.
Chatterjee attributes much of the project’s success to extensive collaboration, which allowed the team to leverage resources across academia, government, and industry, including a partnership with 23andMe. This collaboration facilitated access to large and diverse datasets for evaluating and comparing various polygenic risk score methodologies, including CT-SLEB, on an unprecedented scale.
In the context of safeguarding individual privacy, Chatterjee emphasizes that this partnership with 23andMe was purely for research purposes, with no financial ties between the study authors and the company. He states, “The collaboration with 23andMe was very unique for this project. This partnership gave us access to unprecedentedly large and diverse datasets to evaluate and compare a variety of polygenic risk score methodologies, including CT-SLEB. I don’t think there’s any other paper that has been able to analyze this volume of data before, and that was only possible because of the collaboration with 23andMe.“
Conclusion:
CT-SLEB’s development signifies a crucial step towards bridging the accuracy gap in genetic risk assessments for diverse populations. This innovation not only promises more equitable healthcare but also highlights the significance of addressing biases in algorithms and data representation. The scalability and accessibility of CT-SLEB further reinforce its potential to revolutionize the market for genetic risk assessment tools, making them more inclusive and effective.