- Fusion oncoproteins, arising from chromosomal translocations, are pivotal oncogenic drivers, particularly in pediatric cancers.
- Traditional drug design methods struggle to target these proteins due to their complex structures and lack of distinct binding sites.
- FusOn-pLM, a tailored protein language model developed by Duke University, enhances the representation of fusion oncoproteins, outperforming existing models.
- FusOn-pLM’s training dataset was meticulously curated from FusionPDB and FOdb databases, ensuring comprehensive coverage of fusion oncoprotein sequences.
- The model underwent rigorous evaluation across various benchmark tasks, showcasing superior performance in predicting fusion oncoprotein behaviors and properties.
- Probabilistic masking, particularly the SaLT&PepPr-based approach, significantly enhances FusOn-pLM’s efficacy in discerning protein-protein interaction motifs.
- FusOn-pLM’s advanced embeddings offer unprecedented insights into fusion oncoproteins, paving the way for enhanced therapeutic interventions.
Main AI News:
Fusion oncoproteins, arising from chromosomal translocations, represent pivotal oncogenic drivers, notably in pediatric malignancies. These hybridized proteins present formidable challenges for targeted drug interventions due to their sprawling, disorganized architectures and the absence of discrete binding sites. Conventional drug discovery avenues, such as small molecule approaches, frequently falter owing to their demand for heightened specificity or their tendency to bind vital cellular proteins. Enter protein language models (pLMs), a burgeoning tool offering granular insights into protein functionalities based on sequence analysis. Despite their triumphs across diverse protein landscapes, existing frameworks lack comprehensive training on fusion oncoproteins, thus constraining their utility in therapeutic design for these recalcitrant targets.
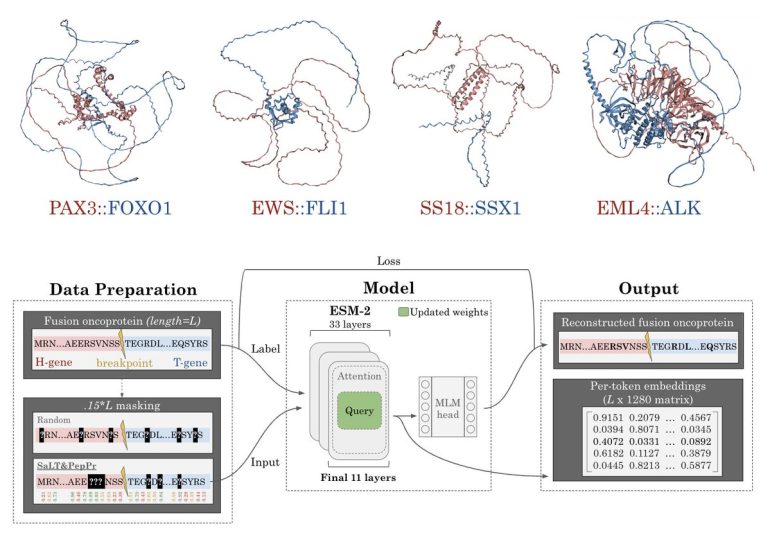
Duke University researchers have pioneered FusOn-pLM, a bespoke protein language model meticulously crafted for fusion oncoproteins. This innovative model refines the state-of-the-art ESM-2 pLM, specifically honing in on fusion oncoprotein sequences sourced from extensive databases. Employing an inventive masked language modeling paradigm, the team zeroes in on critical residues likely implicated in protein interactions. This methodological leap elevates the fidelity of fusion oncoprotein representation, surpassing the foundational ESM-2 model and alternative embedding techniques across diverse evaluative metrics. The enriched embeddings are meticulously engineered to streamline therapeutic interventions targeting these complex proteins. FusOn-pLM stands as an openly accessible resource, poised to catalyze further explorations and applications in this domain.
The FusOn-pLM training dataset underwent rigorous curation leveraging FusionPDB and FOdb repositories, aggregating 41,420 sequences from FusionPDB and 4,536 from FOdb. Selection criteria were stringent, admitting only sequences comprised of the 20 canonical amino acids and not exceeding 2000 amino acids in length to accommodate GPU memory limitations. Following deduplication of overlapping entries, 177 FOdb sequences were earmarked for benchmarking purposes. Subsequent to clustering via the MMSeqs2 tool, employing a minimum sequence identity threshold of 30% and an 80% coverage threshold, clusters were partitioned into training, validation, and testing cohorts in an 80/10/10 ratio. Bespoke benchmarking datasets were curated from FOdb for tasks encompassing the prediction of fusion oncoprotein condensate formation propensity, cellular localization, cancer-related prognostication, and intrinsically disordered region (IDR) analysis.
Evaluation of FusOn-pLM’s efficacy spanned diverse benchmark tasks, encompassing prognostication of fusion oncoprotein phase separation, subcellular localization, and their associations with specific malignancies such as breast invasive carcinoma and stomach adenocarcinoma. A targeted probabilistic masking regime, calibrated to amplify model interpretability, homed in on amino acid residues poised for protein-protein interactions as pinpointed by SaLT&PepPr predictions. This strategic masking, applied to 15% of each sequence, augments the model’s acuity in discerning interaction motifs within fusion oncoproteins. Training iterations involved fine-tuning the advanced ESM-2-650M model by unfreezing the weights and biases of its terminal layers. Benchmarking of FusOn-pLM embeddings vis-à-vis counterparts, including those from ESM-2-650M and manually curated FOdb embeddings, underscored superior performance in IDR prediction and the encapsulation of pivotal physicochemical traits.
FusOn-pLM’s embeddings received a potency boost through probabilistic masking, with the SaLT&PepPr-centric approach yielding optimal results. The model underwent comprehensive evaluations across assorted tasks, showcasing heightened proficiency in forecasting fusion oncoprotein behaviors and properties, including puncta formation propensity and subcellular localization. Moreover, FusOn-pLM distinguished itself in delineating intrinsically disordered regions and their physicochemical attributes, eclipsing alternative embedding methodologies. Visualization methodologies underscored FusOn-pLM’s capacity to demarcate fusion oncoproteins distinctly from constituent elements, underscoring their idiosyncratic attributes and biological significance.
Conclusion:
The development of FusOn-pLM represents a significant leap forward in precision therapy, particularly for targeting fusion oncoproteins. Its superior performance and enhanced capabilities in predicting protein behaviors and properties signify a promising avenue for pharmaceutical companies and research institutions seeking to develop more effective treatments for cancers driven by these complex proteins. Incorporating FusOn-pLM into drug discovery pipelines could accelerate the development of targeted therapies, ultimately improving patient outcomes and reshaping the landscape of cancer treatment.