TL;DR:
- BioPlanner, a collaborative effort between Future House and Oxford, automates the creation of precise scientific protocols for Large Language Models (LLMs) in biology.
- LLMs often struggle with multi-step problem-solving and long-term planning, key aspects of scientific experimentation.
- BioPlanner introduces an automatic evaluation framework and the BIOPROT1 dataset, tailored to enhance LLMs’ planning capabilities in biology and expand to other scientific domains.
- Traditional methods for protocol generation are time-consuming and error-prone.
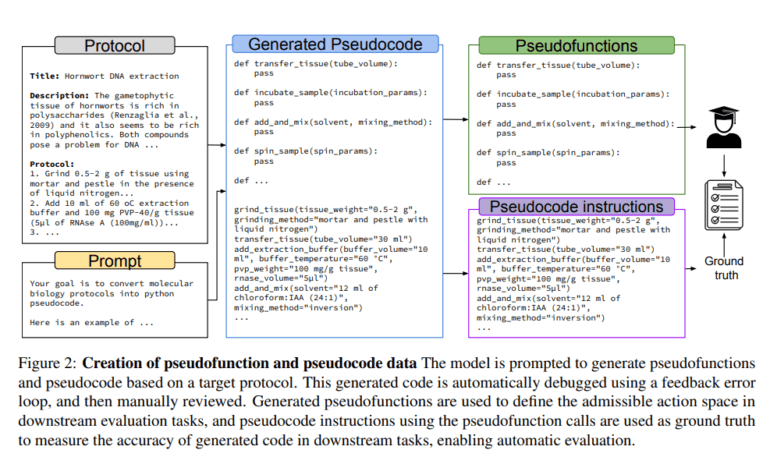
- BIOPROT1 dataset comprises biology protocols translated into pseudocode for clarity.
- BioPlanner teaches LLMs to generate admissible actions and pseudocode, evaluating their ability to reconstruct pseudocode from high-level descriptions.
- GPT-4 is instrumental in converting natural language protocols into pseudocode, simplifying evaluation with unique pseudo functions for each protocol.
- The method improves upon n-gram overlaps and contextual embedding challenges, ensuring robust evaluation metrics.
- GPT-4 outperforms GPT-3.5 in long-term planning and multi-step problem-solving.
- Real-world validation demonstrates BioPlanner’s practical utility in laboratory settings.
Main AI News:
In a pioneering collaboration between Future House and the prestigious University of Oxford, BioPlanner has emerged as a game-changing innovation, designed to address the intricate challenges faced by Large Language Models (LLMs) in the realm of biology. This groundbreaking solution is set to revolutionize the field of scientific experiment planning by automating the generation of precise protocols.
LLMs often struggle with multi-step problems and long-term planning, which are indispensable aspects of scientific experimentation. Recognizing this, the research team, comprised of experts from Align to Innovate, the renowned Francis Crick Institute, Future House, and the esteemed University of Oxford, introduced BioPlanner as the definitive answer to this predicament.
At the heart of this transformative tool lies an automatic evaluation framework and a meticulously curated dataset known as BIOPROT1, specifically tailored to enhance the planning capabilities of LLMs within the domain of biology. With aspirations to expand its applicability across other scientific fields, this development promises to be a game-changer.
The generation of scientific protocols has historically posed significant challenges, characterized by variability in descriptions, sensitivity to minor details, and the imperative need for standardized metrics for evaluation. Traditional methods, marked by their time-consuming nature and susceptibility to errors, were clearly in need of a revolutionary upgrade.
BIOPROT1 emerged as a beacon of innovation, consisting of biology protocols meticulously curated from Protocols.io and translated into pseudocode for clarity and precision. The key to BioPlanner’s effectiveness lies in its ability to instruct LLMs in generating admissible actions and pseudocode for protocols, all while evaluating the LLM’s proficiency in reconstructing pseudocode from high-level descriptions, thus listing admissible pseudocode functions.
At the core of BioPlanner’s functionality is the powerful GPT-4 model, which skillfully transforms natural language protocols into pseudocode, providing a structured representation that simplifies evaluation. The framework defines a unique set of pseudo functions for each protocol, yielding pseudocode and meticulously assessing the model’s capacity to reconstruct it. The research endeavors span a myriad of tasks, including next-step predictions, complete protocol generation, and function retrieval, all accomplished through a combination of shuffled input functions and feedback loops for error identification.
A critical breakthrough achieved through BioPlanner is its ability to tackle the challenges associated with n-gram overlaps and contextual embeddings, thanks to its pseudocode representations. This paves the way for more robust evaluation metrics, ensuring precision and reliability in the assessment of LLMs.
With its unparalleled innovation, BioPlanner presents a solution to the imperative challenge of automating scientific experiment protocols. The method’s effectiveness is incontrovertibly established through rigorous testing on the BIOPROT1 dataset, proving the merits of pseudocode representations. As expected, GPT-4 emerges as the champion, outperforming its predecessor, GPT-3.5, across various tasks, underscoring its superior abilities in long-term planning and multi-step problem-solving.
In a resounding validation of its real-world utility, BioPlanner successfully executed an LLM-generated protocol in a laboratory setting. This practical demonstration underscores the profound impact and relevance of this pioneering method in the realm of scientific experimentation. BioPlanner is poised to lead the way in advancing AI-powered protocol planning, promising a brighter and more efficient future for biology and beyond.
Conclusion:
BioPlanner’s emergence signifies a transformative development in AI-powered protocol planning for LLMs in biology. Its potential to enhance planning abilities, reduce errors, and improve overall efficiency has significant implications for the market, offering advanced solutions for scientific experiment planning in various domains. Businesses and institutions in the research and development sector should closely monitor and consider integrating BioPlanner into their workflow for improved outcomes and efficiency.