TL;DR:
- Genentech and Stanford University collaborate on a revolutionary approach to genetic perturbation experiments.
- Perturb-seq is a technique for understanding gene and cell function by assessing expression responses to genetic disturbances.
- Challenges arise due to the complexity of biological contexts and non-additive genetic interactions.
- Machine learning is leveraged to predict perturbation outcomes, but faces reliability issues due to selection bias.
- A new paradigm combines wet-lab experimentation with an optimal design approach for machine learning model training.
- Active learning principles are introduced but face limitations due to resource constraints.
- ITERPERT, a novel approach, supplements empirical evidence with publicly available prior knowledge sources and employs data fusion techniques.
- ITERPERT demonstrates accuracy comparable to established methods with fewer perturbations.
- The collaboration highlights the power of interdisciplinary research in advancing genetic experimentation precision.
Main AI News:
Genentech and Stanford University, two leading institutions in the field of genetics and cellular research, have joined forces to revolutionize the landscape of genetic perturbation experiments. In a collaborative effort, they have developed an iterative Perturb-seq procedure that leverages the power of machine learning, ushering in a new era of efficiency and precision in the design and execution of perturbation experiments.
The Role of Perturb-seq in Understanding Gene and Cell Function
At the heart of this groundbreaking endeavor lies Perturb-seq, a cutting-edge methodology for conducting pooled genetic screens. This technique relies on assessing the expression response of cells to genetic disturbances, utilizing single-cell RNA sequencing (scRNA-seq) as its cornerstone. Perturb-seq offers a unique window into the intricate world of gene regulation, enabling researchers to engineer cells to specific states and uncover target genes with therapeutic potential.
Challenges in the Perturbation Space
The vast potential of Perturb-seq is met with challenges that arise from its inherent complexity. With an ever-expanding array of biological contexts, cell types, states, and stimuli to consider, the number of required tests grows exponentially. Non-additive genetic interactions further compound the intricacy of these experiments, rendering the direct execution of all experiments impractical, especially when the possibilities number in the billions.
Machine Learning as a Game Changer
Recent research has illuminated a promising path forward by harnessing the power of machine learning. Researchers have developed algorithms that can predict the outcomes of perturbations, individual genes, or gene combinations, using existing Perturb-seq datasets as training data. While these models hold tremendous potential, they are not without their shortcomings. A selection bias introduced during the original experiment’s design has cast a shadow over their reliability.
A Paradigm Shift in Perturb-seq Experiments
Genentech and Stanford University researchers have introduced a novel approach to tackling the perturbation space. In this paradigm, the Perturb-seq assay unfolds in a wet-lab environment, while a machine learning model operates using an interleaving sequential optimal design approach. At each stage of the process, data acquisition and model re-training take place. To ensure the model’s accuracy in predicting unprofiled perturbations, an optimal design technique is employed to select a subset of perturbation experiments strategically. This approach intelligently samples the perturbation space, resulting in a model that thoroughly explores the territory with minimal perturbation experiments.
The Role of Active Learning
Central to this innovative strategy is the concept of active learning, a well-established principle in machine learning. Active learning has found applications in diverse domains, including document classification, medical imaging, and speech recognition. However, successful active learning typically relies on a substantial initial set of labeled examples, a condition that proves challenging in the context of iterative Perturb-seq due to time and budget constraints.
Introducing ITERPERT: A Budget-Friendly Solution
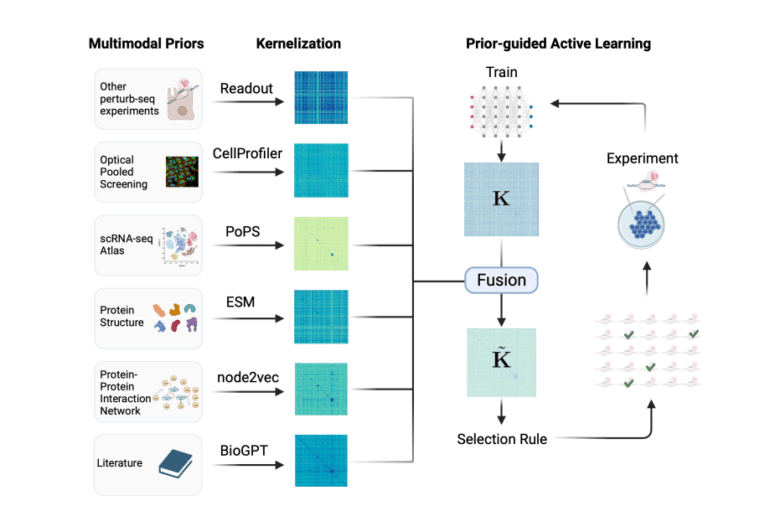
To address the active learning dilemma within the budgetary constraints of Perturb-seq experiments, the research team presents ITERPERT (ITERative PERTurb-seq). Drawing inspiration from data-driven research, this approach emphasizes supplementing empirical evidence with publicly available prior knowledge sources, particularly in the early stages and when resources are limited. These knowledge sources encompass various forms of representation, from networks and text to images and three-dimensional structures.
Harnessing the Power of Knowledge Fusion
To overcome the challenges of utilizing diverse knowledge modalities in active learning, the team employs replicating kernel Hilbert spaces and a kernel fusion approach. This innovative fusion technique merges data from various sources, including physical molecular interactions, Perturb-seq data from comparable systems, and large-scale genetic screens employing different modalities.
A Resounding Success in Empirical Testing
The researchers conducted an extensive empirical investigation using a large-scale single-gene CRISPRi Perturb-seq dataset obtained from a cancer cell line (K562 cells). In a head-to-head comparison with eight established active learning methodologies, ITERPERT demonstrated accuracy levels on par with the top-performing technique while utilizing training data with three times fewer perturbations. Furthermore, when considering batch effects throughout iterations, ITERPERT exhibited exceptional performance in critical gene and genome-scale screens.
Conclusion:
The collaborative efforts of Genentech and Stanford University have yielded a game-changing approach to genetic perturbation experiments. Their innovative techniques, including machine learning and data fusion, not only enhance accuracy but also address resource constraints. This breakthrough paves the way for more efficient and cost-effective genetic research, promising significant advancements in the market for genetic and cellular therapies and applications.