TL;DR:
- Purdue University researchers utilize topological data analysis (TDA) to demystify complex AI prediction models.
- Reeb networks, constructed through TDA, provide a visual representation of prediction strategies in AI models.
- Reeb networks aid in identifying labeling errors in training data and enhance understanding of generalization in image classification.
- Comparison with traditional visualization techniques highlights the superiority of Reeb networks.
- The construction of Reeb networks relies on extensive data, known relationships, and real-valued guides.
- Practical applications include improving product predictions on Amazon and uncovering labeling errors in image datasets.
- Reeb networks also assist in deciphering predictions related to gene mutations, particularly in the BRCA1 gene.
Main AI News:
In the ever-evolving landscape of artificial intelligence and machine learning, complex prediction models have emerged as powerful tools across various scientific domains. However, the intricate web of parameters within these models often shrouds their prediction strategies in obscurity. Enter Purdue University researchers, who have embarked on a mission to demystify these models using innovative topological data analysis (TDA).
In a bid to make sense of the convoluted workings of AI models, including neural networks and machine learning algorithms, Purdue’s team harnessed the potential of TDA to construct Reeb networks—a transformative approach that unveils the topological structure underlying these predictions. This breakthrough methodology promises to shed light on prediction strategies, thereby enhancing our understanding of these complex models.
The Reeb networks introduced by the Purdue researchers essentially transform the abstract prediction landscape into a comprehensible visual format. Within this network, each node signifies a localized simplification of the prediction space, representing clusters of data points with similar predictions. The connections between nodes are forged through shared data points, revealing valuable insights into the relationships between predictions and the underlying training data.
One pivotal application of this innovative approach lies in its capacity to detect labeling errors within training data. The Reeb networks have proven adept at identifying ambiguous regions and prediction boundaries, offering crucial guidance for further exploration and potential error rectification. Moreover, the method has demonstrated its utility in unraveling the intricacies of generalization in image classification and dissecting predictions associated with pathogenic mutations within the BRCA1 gene.
Comparisons with established visualization techniques such as tSNE and UMAP have underscored the superiority of Reeb networks in illuminating the boundaries between predictions and the intricate connections between training data and predictions.
The creation of Reeb networks necessitates certain prerequisites, including an extensive dataset with undisclosed labels, knowledge of relationships among data points, and real-valued guides for predicted values. To construct the Reeb network, the researchers employed a recursive splitting and merging technique known as GTDA (graph-based TDA), showcasing its scalability by analyzing a staggering 1.3 million images from the ImageNet dataset.
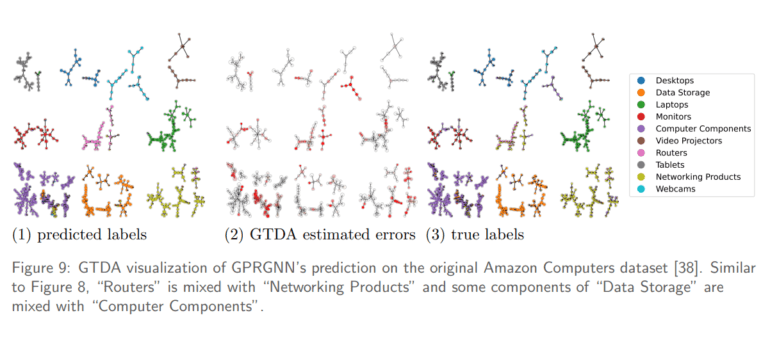
In practical applications, the Reeb network framework was leveraged to enhance the predictive capabilities of a graph neural network on Amazon. By unveiling ambiguities within product categories, it emphasized the imperfections in prediction accuracy, calling for improvements in labeling. Similar insights were gleaned when applying the framework to a pretrained ResNet50 model on the Imagenet dataset, offering a visual taxonomy of images and uncovering the truth behind labeling errors.
The researchers further demonstrated the prowess of Reeb networks in deciphering predictions pertaining to malignant gene mutations, particularly within the BRCA1 gene. These networks spotlighted localized components within the DNA sequence and their correlation with secondary structures, providing invaluable aids for interpretation.
Conclusion:
Purdue’s groundbreaking use of topological data analysis to create Reeb networks represents a significant leap forward in understanding and interpreting complex AI models. This innovation promises to enhance transparency and accuracy in AI applications across various industries, allowing businesses to make more informed decisions and improve the quality of their predictions. It opens up opportunities for refining product recommendations, image classification, and gene mutation analysis, ultimately impacting the market by increasing the reliability and effectiveness of AI-powered solutions.