- ProtEx, developed by Google DeepMind, Google, and University of Cambridge, combines retrieval-based techniques and deep learning for protein function prediction.
- It enhances accuracy, robustness, and generalization to new protein classes by using exemplars from a database.
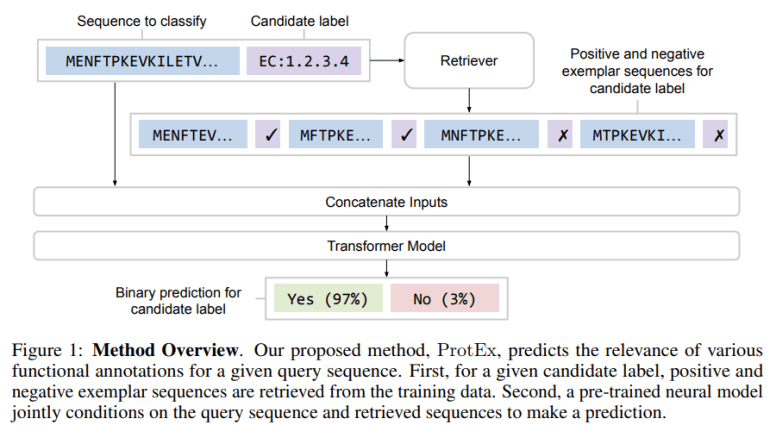
- Inspired by NLP and vision techniques, ProtEx retrieves exemplars using tools like BLAST and trains a neural model to compare them with the query.
- Ablation studies validate ProtEx’s efficacy in pretraining and exemplar conditioning.
- It integrates traditional protein similarity searches with deep learning models, excelling especially with rare and dissimilar sequences.
- ProtEx adapts to new labels without additional fine-tuning, leveraging multi-sequence pretraining.
- Evaluation across multiple datasets shows ProtEx outperforms previous methods in predicting EC numbers, GO terms, and Pfam families.
Main AI News:
As biology delves deeper into the intricate world of proteins, understanding their functions becomes increasingly paramount. Proteins, the workhorses of organisms, play multifaceted roles, and categorizing these roles accurately is no small feat. Enter ProtEx, a cutting-edge solution developed by a collaborative effort from Google DeepMind, Google, and the University of Cambridge. This groundbreaking method combines the prowess of retrieval-based techniques with deep learning to revolutionize protein function prediction.
ProtEx stands out by leveraging exemplars from a vast database, enhancing not only accuracy but also robustness and the ability to generalize to new protein classes. Drawing inspiration from techniques in natural language processing (NLP) and computer vision, ProtEx marries non-parametric similarity searches with deep learning methodologies. By retrieving both positive and negative exemplars using tools like BLAST and training a neural model to compare these with the query, ProtEx achieves state-of-the-art results across various metrics, including predicting Enzyme Commission (EC) numbers, Gene Ontology (GO) terms, and Pfam families.
This innovative approach fills a crucial gap in the field, particularly excelling with rare and dissimilar sequences, often referred to as the “dark matter” of the protein universe. Ablation studies further validate the effectiveness of ProtEx’s pretraining strategy and exemplar conditioning, solidifying its position as a game-changer in protein function prediction.
ProtEx doesn’t reinvent the wheel but rather builds upon existing methods while incorporating novel strategies. Traditional protein similarity searches like BLAST provide the foundation, retrieving homologous sequences to infer functions. However, deep learning models, with their ability to map sequences directly to functions, offer a promising alternative. ProtEx ingeniously integrates these approaches, using BLAST for retrieval and a neural model to refine predictions based on the retrieved exemplars.
What sets ProtEx apart is its adaptability and efficiency. Inspired by retrieval-augmented models in NLP and vision, ProtEx seamlessly incorporates contextual information from retrieved exemplars, enhancing prediction accuracy without the need for extensive fine-tuning. This adaptability extends to new labels, thanks to multi-sequence pretraining, which enables ProtEx to predict protein function labels for a given amino acid sequence effectively.
Evaluation of ProtEx across multiple datasets underscores its prowess. From EC number prediction to GO term classification and Pfam family categorization, ProtEx consistently outperforms previous methods. Whether conditioned on exemplar sequences or handling diverse protein families, ProtEx maintains its stellar performance, setting a new standard in protein function prediction.
Conclusion:
The introduction of ProtEx represents a significant advancement in the protein function prediction market. Its ability to combine retrieval-based techniques with deep learning not only enhances accuracy and robustness but also addresses challenges such as generalization to new protein classes and predicting functions for rare and dissimilar sequences. ProtEx’s performance surpasses previous methods across various classification tasks, indicating its potential to become the go-to solution for protein function prediction in both research and industry settings. This innovation sets a new benchmark, reshaping the landscape of protein analysis and offering promising opportunities for further advancements in the field.