TL;DR:
- Duke University’s biomedical engineers introduce a groundbreaking method for enhancing machine learning efficiency.
- Pairing two machine learning models, one for data collection and the other for analysis, overcomes existing limitations.
- Researchers can leverage this technique to identify and characterize molecules for potential therapeutics.
- Active machine learning’s questioning capability enhances accuracy but faces limitations in complex deep neural networks.
- “Yoked learning” concept, borrowed from education, is applied to machine learning, giving rise to YoDeL.
- YoDeL outperforms or matches active deep learning in identifying molecular characteristics while being significantly faster.
- A provisional patent has been filed for YoDeL, with plans to refine and deploy it in practical applications.
Main AI News:
In a breakthrough development, Duke University’s biomedical engineers have unveiled a groundbreaking method for enhancing the efficiency of machine learning models. This revolutionary approach involves the strategic pairing of two distinct machine learning models, each with a specific role—one to accumulate data and the other to dissect it. The result? A strategy that overcomes the inherent limitations of existing technology while maintaining impeccable accuracy.
This innovative technique promises to usher in a new era for researchers, empowering them to harness the potential of machine learning algorithms to uncover and characterize molecules for potential therapeutic applications and other materials. The implications of this research are documented in the prestigious journal, “Artificial Intelligence in the Life Sciences.”
Traditional machine learning models rely on a fixed dataset provided by researchers to make predictions. Although this approach is generally effective, its limitations stem from the data it is based on, which may lack crucial information or be skewed by an overabundance of one data type, resulting in biased outcomes.
To tackle these limitations, researchers have introduced the concept of active machine learning. In this paradigm, the model possesses the ability to question and seek additional information when gaps in the data are detected. This active approach enhances the model’s accuracy and efficiency compared to its passive counterpart.
However, this promising approach encounters obstacles when applied to complex deep neural networks. These networks, designed to mimic the human brain, demand extensive data and computational power, often exceeding available resources and diminishing their accuracy and efficacy.
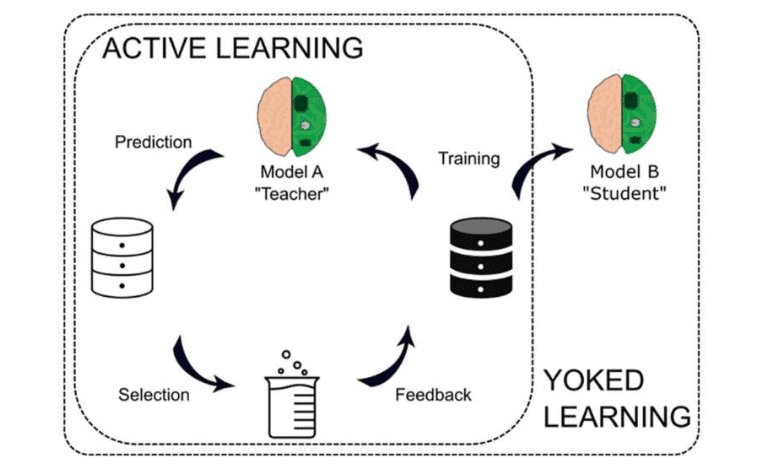
In response to this challenge, Dr. Reker and his team delved into the educational concept of “yoked learning” to potentially transform the landscape of machine learning. Yoked learning involves one student actively learning content, asking questions, and referencing different sources for missing information. Another student is then tasked with reviewing the information highlighted by the first student as crucial for comprehending the lesson.
Dr. Reker was intrigued by the possibility of adapting this concept to machine learning. He explained, “An active machine learning model can navigate through a dataset, identify essential information, and request any missing data it deems significant. We aimed to leverage this active machine learning model to ‘teach’ another model using the data identified as important.”
To assess the efficacy of yoked machine learning versus active machine learning, the team tasked an actively learning model with identifying critical characteristics of molecular compounds, such as potential toxicity and metabolism. They then developed a yoked system, pairing different “teaching” machine learning models with “student” models to identify the same characteristics based on the data selected by the “teaching” model.
While active machine learning outperformed the yoked system in most cases, the team discovered that the teaching model’s performance played a pivotal role in the success of the student models. Just as in traditional education, an ineffective teacher hinders student progress. If the teaching model failed to identify valuable data, the student model struggled to decipher it.
These findings inspired Dr. Reker and his team to explore yoked learning further, particularly with a deep neural network model as the “student.” This novel approach, named Yoked Deep Learning (YoDeL), employed an active machine learning algorithm as the “teacher” to guide data acquisition for the deep neural network “student.”
In comparative studies encompassing various models, YoDeL consistently outperformed or matched the accuracy of active deep learning systems in identifying molecular characteristics. Moreover, YoDeL demonstrated remarkable speed, often completing tasks in just minutes, whereas deep active learning required hours or even days.
The team has already filed a provisional patent for the YoDeL technique and has ambitious plans to refine its parameters and deploy it in practical applications. Dr. Reker expressed optimism about the future, stating, “YoDeL’s ability to harness the strengths of classical machine learning models to enhance the efficacy of deep neural networks makes it an exciting tool for an ever-evolving field. We believe that YoDeL will soon play a crucial role in discovering new drugs and drug delivery solutions.”
Conclusion:
The development of YoDeL marks a significant advancement in machine learning for molecular discovery. Its ability to combine the strengths of classical machine learning models with the efficiency of deep neural networks positions it as a game-changer in drug discovery and material characterization. This innovation has the potential to streamline research processes, reduce costs, and accelerate the development of new drugs and drug delivery solutions, making it a valuable asset for the evolving market of pharmaceuticals and materials science.