TL;DR:
- Scientists from Harvard and MIT developed BioAutoMATED, an automated machine learning (autoML) platform.
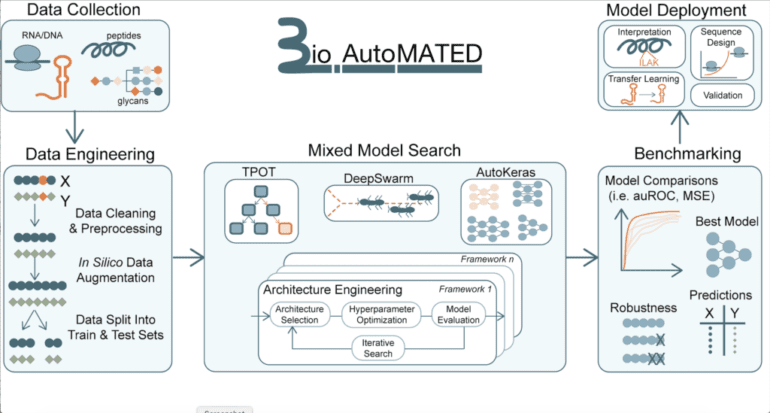
- BioAutoMATED simplifies ML for biologists, eliminating technical barriers and reducing coding requirements.
- It accommodates various ML models and targets biological sequences for research.
- The platform outperformed other autoML tools and some expert-built models in under 30 minutes.
- BioAutoMATED’s versatile features include scrambled control tests, data saturation tests, and interpretation results.
- It offers actionable insights and optimized de novo design sequences for experiments.
- The platform is user-friendly, enabling researchers to access ML capabilities with only ten lines of input code.
- BioAutoMATED aims to democratize machine learning for biologists with limited ML expertise.
Main AI News:
In the rapidly evolving landscape of biotechnology, automated machine learning (autoML) emerges as a game-changer, empowering biologists to surmount technical hurdles and harness the power of computational models. At the forefront of this revolution are scientists from the Wyss Institute for Biologically Inspired Engineering at Harvard University and MIT. Driven by the belief that simplicity is the key to progress, they are propelling autoML to the forefront of biological research.
For graduate student Jackie Valeri, the potential of autoML to address real-world challenges is awe-inspiring. By facilitating the transfer of data to training algorithms and autonomously searching for the most fitting ML architecture, autoML nullifies the need for extensive computational expertise that has long hindered progress. In a landmark paper published in Cell Systems, Valeri and her colleagues unveiled their groundbreaking BioAutoMATED platform, designed to embrace various ML models and cater to biological sequences. This revolutionary tool opens doors for systems and synthetic biologists, even those with limited ML experience, to partake in the scientific data revolution.
What sets BioAutoMATED apart from other autoML tools is its versatility. By integrating three existing AutoML tools—AutoKeras, DeepSwarm, and TPOT—the platform crafts an ideal model tailored to the user’s dataset. The standardized output is presented as a set of folders, each associated with a unique search technique, displaying the best-performing model in both graphic and text file formats.
The beauty of BioAutoMATED lies in its self-learning capabilities. Model selection, an intricate task requiring specialized expertise, becomes accessible to biologists who might lack computational prowess. No longer must they rely on ML specialists, as BioAutoMATED paves the way for autonomous research tailored to the biologist’s domain knowledge.
Until now, the excitement around machine learning in biology was often hampered by the complexity of coding. ML models demanded extensive lines of code, acting as a daunting barrier for aspiring researchers. However, with BioAutoMATED’s user-friendly approach, the installation of Docker and a mere ten lines of input code offer access to unparalleled data analysis, making the platform a true “quick start” for researchers worldwide.
A testament to its efficacy, BioAutoMATED has outperformed both other autoML tools and certain models crafted by professional ML experts. Notably, it accomplishes this feat in under 30 minutes, revolutionizing the speed of data-driven research.
With features like scrambled control tests, data saturation tests, interpretation results, and design outcomes, BioAutoMATED empowers biologists to identify crucial insights, leading to groundbreaking hypotheses and experiments. From deciphering sequence logos to optimizing ribosome binding sites and classifying glycans, the platform’s impact extends across a multitude of biological domains.
Wyss postdoctoral fellow Luis Soenksen predicts a future where BioAutoMATED collaborates harmoniously with ChatGPT, collectively presenting researchers with optimal questions, data, and ML models. As adoption grows, BioAutoMATED’s impact will undoubtedly expand, urging the team to enhance its user interface for seamless accessibility.
While BioAutoMATED simplifies the ML process, Valeri acknowledges that machine learning experts may still achieve finer-tuned results. Nevertheless, the platform’s mission is clear—to empower researchers with limited ML expertise, democratizing the realm of machine learning for the greater scientific good.
Conclusion:
The BioAutoMATED breakthrough marks a significant advancement in the field of bioinformatics. By simplifying the application of machine learning in biological research, this platform eliminates technical barriers and empowers biologists to explore complex datasets with ease. As BioAutoMATED gains traction and enhances its user interface, it has the potential to revolutionize the market by democratizing machine learning capabilities and accelerating discoveries in the biological sciences. Biotechnology companies and research institutions should closely monitor this development, as it promises to shape the future of data-driven biological research and unlock new opportunities for innovation and breakthroughs in the market.