TL;DR:
- ProtHyena, developed by the Tokyo Institute of Technology, introduces the Hyena operator for analyzing protein data.
- It excels in capturing both long-range context and single amino acid resolution in protein sequences.
- The model is pretrained on the Pfam dataset and fine-tuned for various protein-related tasks.
- ProtHyena outperforms traditional language models based on the Transformer and BERT architectures.
- It achieves state-of-the-art results in tasks such as Remote Homology and Fluorescence prediction.
- The Hyena operator and its natural protein vocabulary set ProtHyena apart from the competition.
- The model demonstrates potential in Secondary Structure Prediction (SSP) and Stability tasks.
Main AI News:
In the ever-evolving landscape of biological research, the importance of proteins cannot be overstated. They serve as the building blocks of life, providing the essential amino acids needed for various cellular functions. Unlocking the mysteries of proteins has long been a central pursuit in the realm of human biology and health. To this end, advanced machine-learning models for protein representation have emerged as a beacon of hope.
Self-supervised pre-training, inspired by the strides made in natural language processing, has ushered in a new era of protein sequence representation. It has undeniably improved our understanding of these biological workhorses. However, the road to comprehending proteins in their entirety is paved with challenges, particularly when it comes to handling longer sequences and maintaining contextual understanding.
Linearized and sparse approximations have been deployed to alleviate the computational demands imposed by these complex molecules. Still, these strategies often exact a toll on the expressivity of the models. Even the most formidable models, boasting over 100 million parameters, struggle to grapple with the intricacies of larger inputs. The intricate dance of individual amino acids adds a layer of complexity that demands a nuanced approach for accurate modeling.
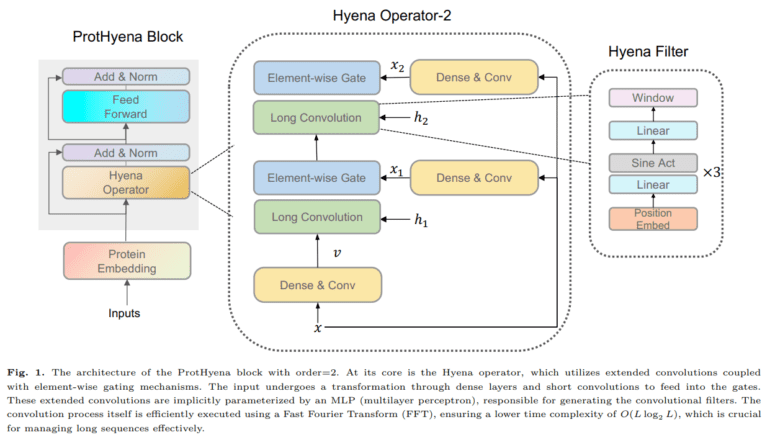
Enter ProtHyena, the brainchild of researchers from the Tokyo Institute of Technology, Japan. This groundbreaking protein language model represents a monumental leap forward. ProtHyena introduces the Hyena operator, a novel approach to analyzing protein data. In stark contrast to traditional attention-based methods, ProtHyena is purpose-built to embrace both long-range context and the granular, single amino acid resolution in real protein sequences.
The journey of ProtHyena begins with self-supervised pre-training on the vast Pfam dataset. From there, the model embarks on a rigorous journey of fine-tuning, honing its prowess for a myriad of protein-related tasks. In some instances, its performance rivals and even surpasses the most state-of-the-art approaches.
Conventional language models, grounded in the Transformer and BERT architectures, have demonstrated their mettle across diverse applications. Yet, they grapple with the quadratic computational complexity of the attention mechanism. This constraint hinders their efficiency and curtails the length of context they can effectively process. Enter a host of innovations, such as factorized self-attention and the Performer, which seek to address the computational cost of self-attention for longer sequences. However, these solutions often necessitate a compromise on model expressivity.
ProtHyena emerges as a trailblazer by harnessing the power of the Hyena operator to surmount the limitations that have plagued traditional language models. By treating each amino acid as an individual token within the natural protein vocabulary, ProtHyena ushers in a new era of representation. Special character tokens for padding, separation, and unknown characters further enhance its flexibility.
The Hyena operator, defined by a recurrent structure featuring long convolutions and element-wise gating, sets ProtHyena apart from the crowd. Moreover, the study extends its reach by comparing ProtHyena with a variant model called ProtHyena-bpe. The latter employs byte pair encoding (BPE) for data compression and embraces a larger vocabulary size.
The results are nothing short of remarkable. ProtHyena shatters the limitations that have held back traditional models based on the Transformer and BERT architectures. Across various downstream tasks, from Remote Homology to Fluorescence prediction, ProtHyena emerges as the undisputed champion, outperforming contemporary models like TAPE Transformer and SPRoBERTa.
When it comes to Remote Homology, ProtHyena delivers an astounding accuracy rate of 0.317, leaving other models trailing behind at 0.210 and 0.230. For Fluorescence prediction, ProtHyena demonstrates its mettle with a Spearman’s r of 0.678, underscoring its ability to grasp the intricacies of complex protein properties. Even in Secondary Structure Prediction (SSP) and Stability tasks, ProtHyena showcases its immense potential, even though the specifics of these achievements warrant further exploration.
Conclusion:
ProtHyena stands as a testament to the relentless pursuit of excellence in the field of protein representation. With its innovative Hyena operator, it has shattered the boundaries that have long constrained traditional language models. As we embark on a new era of biological discovery, ProtHyena paves the way for a deeper understanding of proteins and their critical role in our biological tapestry.