- SleepFM, a multi-modal foundation model for sleep analysis, is introduced by researchers at Stanford University and the Technical University of Denmark.
- It leverages a vast dataset of over 14,000 participants’ sleep recordings, totaling more than 100,000 hours of data collected between 1999 and 2020.
- SleepFM utilizes a contrastive learning approach to integrate brain activity, ECG, and respiratory signals, significantly enhancing the accuracy of sleep analysis.
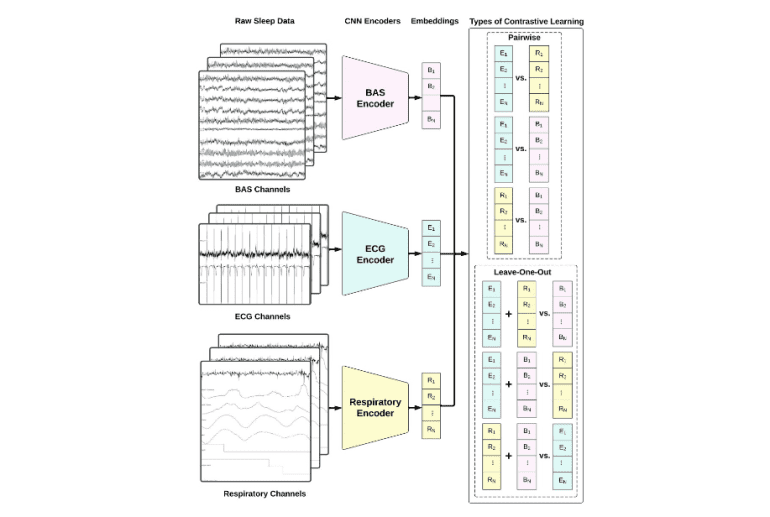
- The model employs three 1D convolutional neural networks (CNNs) tailored to handle specific characteristics of each modality.
- SleepFM outperforms end-to-end CNNs in sleep stage classification and sleep-disordered breathing detection, showcasing superior accuracy and efficiency.
- It achieves remarkable accuracy in demographic attribute classification, accurately predicting age and gender from short physiological data clips.
Main AI News:
The realm of sleep medicine stands as a pivotal domain, necessitating continuous advancements in monitoring and deciphering physiological cues to diagnose sleep-related ailments and fathom sleep behaviors. Techniques like polysomnography (PSG) play a crucial role by tracking brain, cardiac, and respiratory activities during sleep, furnishing a meticulous insight into an individual’s sleep well-being. These signals serve as linchpins in delineating sleep stages and unearthing sleep disorders. A typical PSG setup encompasses an array of modalities such as electroencephalograms (EEG), electrooculograms (EOG), electromyograms (EMG), electrocardiograms (ECG), and respiratory channels. Each modality offers a distinct lens: brain activity signals (BAS) gauge cerebral function, ECG traces heart rhythms, while respiratory sensors quantify breathing patterns, collectively presenting a holistic evaluation of sleep health.
The imperative task of accurately scrutinizing sleep data arises from the labyrinthine nature of sleep disorders. Manual analysis, entailing visual scrutiny by adept technicians, proves to be time-intensive, laborious, and susceptible to inaccuracies. This conventional approach grapples with substantial impediments, particularly amidst the burgeoning influx of sleep data. Consequently, there exists an exigent call for automated methodologies adept at efficiently and precisely dissecting sleep data across myriad physiological channels. The overarching aim is to engender robust models capable of navigating the intricacies of sleep data and furnishing dependable diagnoses.
Current methodologies for sleep data analysis predominantly lean on supervised deep learning paradigms. While these models exhibit promise in automating sleep staging and classifying sleep disorders like sleep-disordered breathing (SDB), they predominantly hinge on annotated data from specific tasks and often fail to harness the full spectrum of physiological signals available from PSG. For instance, deep learning models like convolutional neural networks (CNNs) and recurrent neural networks (RNNs) have been advocated for sleep-scoring endeavors but frequently lag in terms of generalizability and resilience. Moreover, despite the success of contrastive learning (CL) in diverse domains, its application in amalgamating BAS, ECG, and respiratory signals for sleep analysis remains largely untapped.
In a groundbreaking development, researchers from Stanford University and the Technical University of Denmark have unveiled SleepFM, an innovative multi-modal foundation model tailored for sleep analysis. This model capitalizes on an extensive repository of multi-modal sleep recordings encompassing over 14,000 subjects, amassing a staggering 100,000 hours of sleep data amassed between 1999 and 2020 at the Stanford Sleep Clinic. SleepFM adopts a contrastive learning framework to seamlessly integrate brain activity, ECG, and respiratory signals, thereby enabling the model to encapsulate comprehensive physiological portrayals and markedly augmenting the precision of sleep analysis.
SleepFM harnesses three 1D convolutional neural networks (CNNs) to derive embeddings from each modality (BAS, ECG, and respiratory signals). The architectural blueprint of these models is underpinned by a 1D CNN architecture tailored for classifying ECG readings. Each CNN is meticulously calibrated to grapple with the idiosyncrasies of its corresponding modality: 10 channels for BAS, 2 for ECG, and 7 for respiratory channels. Introducing a pioneering leave-one-out contrastive learning methodology yields superior outcomes vis-à-vis the conventional pairwise contrastive learning approach in encapsulating the symbiotic interplay between diverse physiological signals.
In the realm of sleep stage classification, SleepFM notches up a macro AUROC of 0.88 and a macro AUPRC of 0.72, juxtaposed against 0.72 and 0.48 by end-to-end CNNs. For the detection of sleep-disordered breathing, SleepFM eclipses CNNs with an AUROC of 0.85 and an AUPRC of 0.77, surpassing 0.69 and 0.61 by CNNs. Furthermore, SleepFM’s embeddings exhibit a commendable 48% top-1 average accuracy in retrieving corresponding recording snippets from other modalities amidst a pool of 90,000 contenders. These findings underscore the model’s prowess in harmonizing diverse physiological signals and elevating the accuracy and efficacy of sleep analysis.
The triumph of the model can largely be attributed to its adeptness in imbibing rich, multi-modal representations of physiological data, pivotal for precise sleep analysis. SleepFM also shines in demographic attribute classification, showcasing remarkable accuracy in prognosticating age and gender from succinct 30-second clips of physiological data. The model achieves AUROCs of 0.982, 0.852, 0.784, and 0.915 across the age cohorts 0-18, 18-35, 35-50, and 50+, respectively. As for gender classification, the AUROC scales to 0.850, significantly outperforming rudimentary models.
Conclusion:
The introduction of SleepFM marks a significant advancement in the field of sleep analysis, offering a comprehensive and efficient solution for diagnosing sleep disorders and understanding sleep patterns. Its integration of diverse physiological signals and superior performance compared to existing models signifies promising opportunities for improved healthcare outcomes and enhanced patient care in the market.