TL;DR:
- Traditional CNNs struggle with global information capture in 3D medical image segmentation.
- Transformer-based models like TransBTS and UNETR improve global insights but face computational challenges.
- Mamba, a state space model, addresses long-range sequence modeling efficiently.
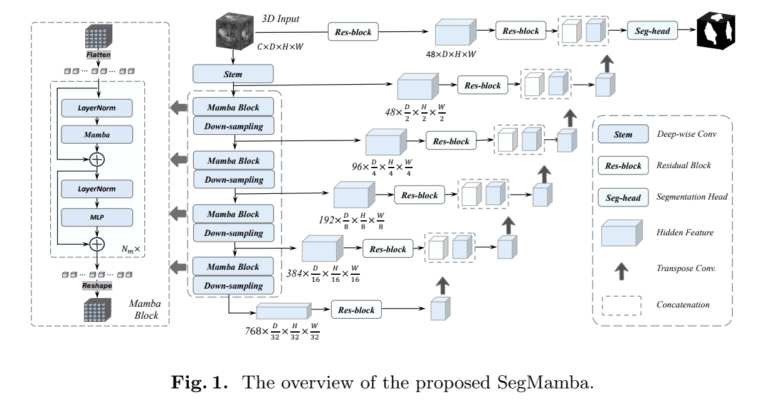
- SegMamba, combining a U-shape structure with Mamba, excels in whole-volume global feature modeling.
- Extensive experiments affirm SegMamba’s effectiveness and superior processing speed.
Main AI News:
In the realm of 3D medical image segmentation, the quest for more effective models never ceases. Traditionally, convolutional neural networks (CNNs) have faced a daunting challenge in grasping global information from high-resolution 3D medical images. One innovative approach involves employing depth-wise convolution with larger kernel sizes, broadening the spectrum of features captured. However, the need to establish relationships across distant pixels remains a hurdle for CNN-based techniques.
The emergence of transformer architectures has paved the way for substantial progress in this domain. Transformer-based models, like TransBTS and UNETR, have harnessed self-attention mechanisms to extract global insights for 3D medical image segmentation. TransBTS ingeniously combines 3D-CNN with transformers, while UNETR adopts the Vision Transformer (ViT) for contextual understanding. Nevertheless, transformer-based methods grapple with computational challenges arising from the high resolution of 3D medical images, leading to reduced processing speed.
In response to the demand for long-range sequence modeling, the introduction of Mamba, a state space model (SSM), proved to be a game-changer. Mamba efficiently models long-range dependencies through a selection mechanism and a hardware-aware algorithm, making it invaluable in computer vision tasks. U-Mamba, for example, has successfully integrated the Mamba layer to enhance medical image segmentation. Simultaneously, Vision Mamba introduces the Vim block, incorporating bidirectional SSM for comprehensive global context modeling and position embeddings for location-aware insights. VMamba further introduces a CSM module, bridging the gap between 1-D array scanning and 2-D plain traversing. Nonetheless, traditional transformer blocks grapple with the challenge of handling large-size features, necessitating the modeling of correlations within high-dimensional features for enhanced visual understanding.
Inspired by these advancements, researchers at the Beijing Academy of Artificial Intelligence introduced SegMamba. This novel architecture seamlessly fuses the U-shape structure with Mamba, enabling the modeling of whole-volume global features across various scales. SegMamba leverages Mamba’s capabilities specifically for 3D medical image segmentation, offering an exceptional solution to the challenges faced by conventional CNN and transformer-based methods.
Extensive experimentation on the BraTS2023 dataset has solidified SegMamba’s reputation for effectiveness and efficiency in 3D medical image segmentation tasks. In contrast to Transformer-based techniques, SegMamba relies on the principles of state space modeling to excel in capturing whole-volume features while maintaining superior processing speed. Even when dealing with volumetric features at a resolution of 64 × 64 × 64 (equivalent to a sequential length of approximately 260k), SegMamba shines as a beacon of efficiency.
Conclusion:
SegMamba’s introduction signifies a significant advancement in the 3D medical image segmentation market. Its capacity to efficiently capture long-range dependencies while maintaining outstanding processing speed promises enhanced precision and productivity in medical imaging applications. This innovation is poised to reshape the market landscape by providing a transformative solution to the challenges faced by existing models.