TL;DR:
- Foundational AI models like scGPT and Geneformer explored in single-cell biology.
- Zero-shot performance was assessed in a rigorous study.
- Both models performed below baseline for key metrics.
- The variability observed in Geneformer across different datasets.
- The pretraining dataset impacted scGPT’s performance positively.
- Models struggled with batch effect correction.
- Geneformer outperformed scGPT in gene expression reconstruction.
- Market implications are discussed in the conclusion.
Main AI News:
In the realm of single-cell biology, the utilization of foundational models has emerged as a significant point of discourse within the research community. Notably, models such as scGPT, GeneCompass, and Geneformer have emerged as promising tools in this domain. However, the effectiveness of these models, particularly in zero-shot scenarios where exploratory experiments and a dearth of well-defined labels are prevalent, has raised questions. This article delves into this very issue, conducting a meticulous evaluation of the zero-shot capabilities of these models.
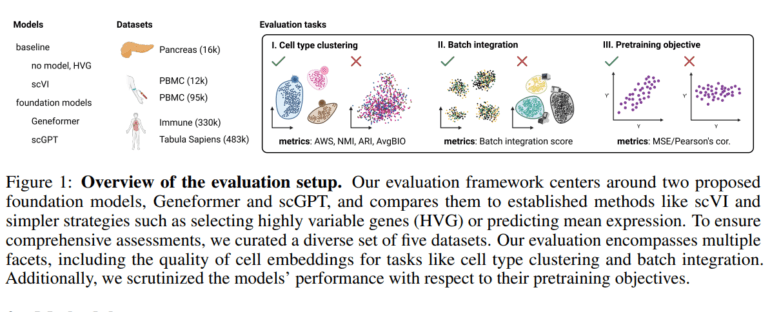
Historically, researchers have relied on fine-tuning these models for specific tasks. Yet, the limitations of such an approach become conspicuous when applied to the intricacies of single-cell biology, coupled with the formidable computational demands it entails. Consequently, in response to this challenge, a team of researchers at Microsoft embarked on an extensive assessment of Geneformer and scGPT’s foundational models across various dimensions, encompassing diverse datasets and a spectrum of tasks, including the utility of embeddings for cell type clustering, batch effect correction, and the models’ proficiency in input reconstruction, all rooted in their pretraining objectives.
The rationale behind selecting these two models stems from the availability of their pretrained weights at the time of assessment. To conduct their evaluation, the researchers harnessed five distinct human tissue datasets, each posing distinct and pertinent challenges to the field of single-cell analysis. To facilitate comparisons, the researchers introduced scVI, a generative model trained on each dataset, into the mix. Several metrics were employed to gauge the models’ performance across these tasks:
- Cell Embeddings Evaluation: The Average Silhouette Width (ASW) and Average Bio (AvgBIO) scores were employed to measure the uniqueness of cell types within the embedding space.
- Batch Integration: A variant of the AWS score, ranging from 0 to 1, was utilized to assess the extent of batch separation, with 0 indicating complete segregation and 1 indicating perfect batch mixing.
- Pretraining Objective Performance: Mean Squared Error (MSE) and Pearson’s correlation were adopted to evaluate the performance of scGPT and Geneformer in their pretraining objectives.
Disappointingly, both scGPT and Geneformer fell short of the baseline strategies for both metrics. Geneformer exhibited notable variability across different datasets, while scGPT, although surpassing the base model scVI for one dataset, lagged behind for two others. Subsequently, the researchers turned their attention to the impact of the pretraining dataset on model performance, with a primary focus on scGPT. Notably, all variants of scGPT demonstrated improved median scores.
When subjected to the assessment of batch effects, both models yielded lackluster results, frequently trailing behind models like scVI. This suggests that these models may not possess complete robustness when it comes to handling batch effects in zero-shot scenarios. In the final round of evaluations, it was revealed that scGPT struggled to reconstruct gene expressions, while Geneformer displayed a more favorable performance. In a head-to-head comparison against a baseline, the baseline predictions consistently outperformed all variants of scGPT, with Geneformer surpassing the average rankings in one of the datasets.
Conclusion:
The evaluation reveals that scGPT and Geneformer face challenges in zero-shot single-cell biology tasks. While they show promise, their performance falls short of expectations, especially in handling batch effects and gene expression reconstruction. This suggests a need for further refinement to meet the specific demands of the market, emphasizing the importance of continued development in this evolving field.