- Researchers utilize machine learning to analyze over 100,000 genetic sequences in human brain cells.
- Identified 150+ variants potentially causing neurological disorders.
- Study led by Gladstone Institutes and UCSF provides insights into brain development and disorders.
- Organoids prove effective in replicating genetic variants found in human brain tissue.
- Machine learning enables predictive analysis of gene regulatory activity, accelerating research outcomes.
Main AI News:
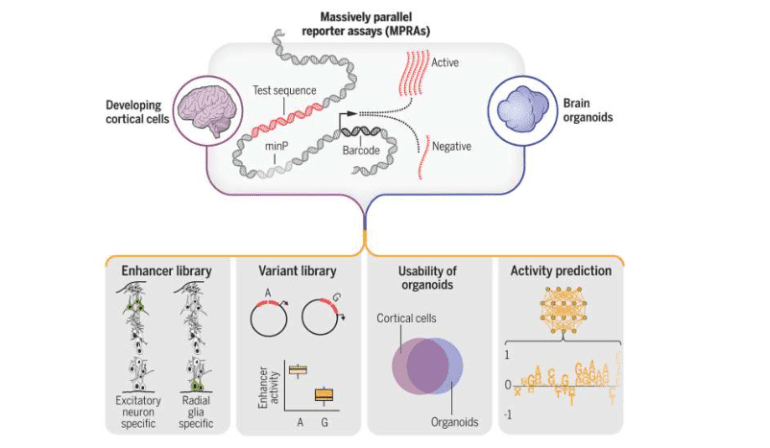
In a groundbreaking scientific endeavor enriching our comprehension of genetic dynamics influencing brain maturation and predisposing individuals to neuropsychiatric ailments, a consortium of researchers amalgamated high-throughput assays with machine learning algorithms to scrutinize over 100,000 genetic sequences within human brain cells, unveiling more than 150 mutations likely implicated in disease etiology.
This pivotal research, spearheaded by scholars from Gladstone Institutes and the University of California, San Francisco (UCSF), has yielded a comprehensive compendium of genetic sequences orchestrating brain evolution, heralding the advent of novel diagnostic modalities and therapeutic avenues for neurological afflictions such as schizophrenia and autism spectrum disorders. Dubbed “Massively Parallel Characterization of Regulatory Elements in the Developing Human Cortex,” the study is featured in the esteemed journal Science.
“We amassed an extensive dataset from noncoding regions of DNA, presumed to exert profound influence on brain development or pathology,” elucidates Senior Investigator Katie Pollard, Ph.D., also assuming the role of director at the Gladstone Institute for Data Science and Biotechnology. “Through functional interrogation of over 100,000 sequences, we discerned their impact on gene expression, pinpointing sequence variations potentially modulating their activity in disease states.”
Pollard, in tandem with Nadav Ahituv, Ph.D., a professor in the Department of Bioengineering and Therapeutic Sciences at UCSF, and director of the UCSF Institute for Human Genetics, helmed this ambitious initiative. The experimental endeavors, particularly focused on brain tissue, were steered by Tomasz Nowakowski, Ph.D., an associate professor of neurological surgery in the UCSF Department of Medicine.
In total, the consortium unearthed 164 mutations associated with psychiatric disorders and identified 46,802 sequences harboring enhancer attributes in nascent neurons, indicative of their pivotal role in gene regulation.
“These enhancers hold promise for mitigating psychiatric maladies arising from dysfunctional gene expression,” remarks Ahituv. “Given that myriad disorders stem from aberrant gene function, harnessing these enhancers could potentially rectify gene deficiencies.”
Pioneering Insights from Organoids and Machine Learning
In addition to delineating enhancers and disease-associated sequences, the study proffers pioneering insights in two cardinal domains.
Firstly, researchers replicated segments of their investigation employing brain organoids derived from human stem cells, demonstrating their efficacy as surrogate models. Notably, the majority of genetic aberrations detected in human brain tissue were mirrored in the cerebral organoid.
“Our organoid exhibited remarkable fidelity to the human brain,” asserts Ahituv. “As we embark on exploring additional sequences implicated in neurodevelopmental disorders, the organoid emerges as a robust platform for deciphering gene regulatory dynamics.“
Secondly, through assimilating copious DNA sequence data and gene regulatory profiles into a machine learning framework, the team successfully trained the algorithm to prognosticate the activity of specific sequences. This paradigm facilitates “in-silico” experiments, empowering researchers to anticipate experimental outcomes preemptively, thus expediting discoveries and conserving resources, particularly in scenarios necessitating extensive biological datasets.
Sean Whalen, Ph.D., a senior research scientist in the Pollard Lab at Gladstone and a co-first author of the study, underscores the efficacy of the machine learning model in extrapolating gene expression patterns based on noncoding DNA regions.
“The model’s proficiency in predicting outcomes, devoid of prior exposure to the dataset, underscores its adeptness in discerning fundamental principles governing gene regulation in developing brain cells,” elucidates Whalen. “This paradigm holds promise in envisaging the combinatorial effects of genetic variants, ushering in a new era of research possibilities.”
Charting a New Trajectory for Brain Research
This seminal investigation culminated within the ambit of the PsychENCODE Consortium, an interdisciplinary collaboration endeavoring to amass comprehensive gene expression and regulatory datasets from human brains across diverse psychiatric disorders and developmental stages.
By disseminating multiple studies, the consortium endeavors to illuminate the intricacies of enigmatic psychiatric conditions, spanning autism to bipolar disorder, while catalyzing novel therapeutic modalities.
“Our research augments this burgeoning repository of knowledge, underscoring the efficacy of human cells, organoids, functional screening modalities, and deep learning in unraveling regulatory elements and variants governing human brain development,” affirms Chengyu Deng, Ph.D., a postdoctoral researcher at UCSF and a co-first author of the study.
Conclusion:
The breakthrough in decoding genetic influence on brain development through machine learning signifies a paradigm shift in neuroscience research. With the ability to predict gene regulatory activity and identify disease-associated variants, this advancement holds promise for the development of targeted diagnostics and therapeutics in the neurological disorder market. Pharmaceutical companies and biotech firms may leverage these insights to drive innovation and address unmet medical needs in psychiatric care.