- Protein engineering, propelled by machine learning (ML), is poised to transform industries like antibodies, drugs, and agriculture.
- ML-driven approaches accelerate protein modification by analyzing vast datasets and predicting mutation impacts.
- Advanced mathematical tools like topological data analysis (TDA) and NLP models enhance data representation and model training.
- Innovations include sequence-driven deep protein language models and structure-based TDA models.
- Challenges persist in data preprocessing and iterative optimization, but future directions aim to address these issues.
Main AI News:
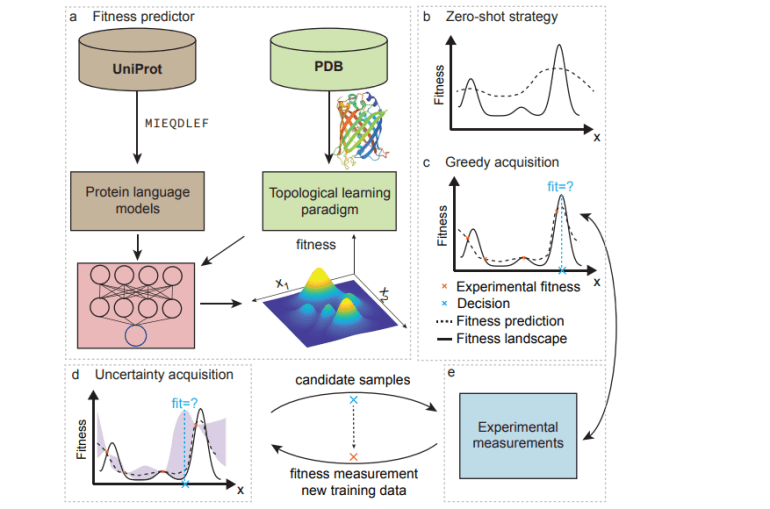
The domain of protein engineering, a swiftly progressing sector within biotechnology, stands poised to reshape numerous industries, spanning antibody innovation, pharmaceutical exploration, agricultural sustainability, and environmental conservation. Conventional methodologies such as directed evolution and rational design have proven pivotal. Nonetheless, the sheer expansiveness of the mutational landscape renders these approaches cost-prohibitive, time-intensive, and somewhat constrained in their applicability. The harnessing of extensive protein repositories coupled with sophisticated machine learning (ML) architectures, particularly those inspired by natural language processing (NLP), has markedly expedited protein engineering endeavors. Innovations in topological data analysis (TDA) alongside AI-driven tools for protein structure prognostication, exemplified by breakthroughs like AlphaFold2, have further augmented the efficacy of ML-driven strategies in protein engineering.
Machine learning-facilitated protein engineering (MLPE) capitalizes on data-centric methodologies to bolster the efficiency and efficacy of protein modification endeavors. ML algorithms adeptly generate and evaluate a plethora of protein variants, discerning the ramifications of mutations and optimizing the protein-fitness landscape, even with sparse experimental data. MLPE entails a holistic framework encompassing data aggregation, feature extraction, model refinement, and iterative validation, synergized with high-throughput sequencing and screening technologies.
Advanced mathematical frameworks such as TDA and NLP-powered models assume pivotal roles in data representation, pivotal for precise model training and prognostication. Notwithstanding notable strides, persistent challenges including data preprocessing intricacies, feature extraction nuances, and iterative optimization hurdles persist. This discourse delves into these challenges while delineating prospective trajectories in the domain, aimed at augmenting the methodologies and outcomes of MLPE endeavors.
Sequence-Driven Deep Protein Language Models
Innovations in NLP have engendered computational paradigms for scrutinizing protein sequences, treating them analogously to linguistic constructs. Sequence-driven protein language models, leveraging local evolutionary insights from homologs and global datasets from expansive protein repositories such as UniProt, have emerged to prognosticate proteins’ structural and functional attributes. Methodologies span from localized models employing Hidden Markov Models (HMMs) and variational autoencoders (VAEs) to overarching frameworks deploying formidable NLP architectures akin to Transformers. Hybrid strategies, entailing the fine-tuning of global models with localized data, further bolster prognostic accuracy, exemplified by pioneering models like eUniRep and Transcription.
Topology-Driven Structural Data Analysis (TDA) Models
Structural models predicated on TDA circumvent the constraints of sequence-driven models by assimilating stereochemical insights. TDA, grounded in algebraic topology, elucidates intricate geometric data and unveils topological constructs. Persistent homology, a seminal TDA methodology, scrutinizes multiscale data, while persistent cohomology and element-specific persistent homology (ESPH) amplify this by accommodating heterogeneous data. Persistent topological Laplacians additionally capture data intricacies. Graph neural networks (GNNs) and topological deep learning amalgamate connectivity and shape insights, propelling protein structure scrutiny and function prognostication with ramifications extending to drug discovery and protein engineering realms.
AI-Assisted Protein Engineering: Predicaments and Resolutions
Protein engineering embodies a multifaceted optimization conundrum, striving to pinpoint the quintessential amino acid sequence that optimizes specific attributes such as functionality, stability, and selectivity. This conundrum is exacerbated by the sheer expanse of sequence space and the intricate, epistatic nature of the fitness landscape, where amino acid interactions exhibit high interdependency and nonlinearity. Conventional methodologies like directed evolution often stumble upon local optima and necessitate assistance in navigating the multidimensional fitness terrain. Furthermore, experimental methodologies are encumbered by the profusion of potential mutations and the throughput constraints of assays, rendering exhaustive exploration of the entire sequence space impracticable.
Recent strides in machine learning have markedly refined the protein engineering paradigm by facilitating expedited exploration and optimization within this expansive search space. Machine learning frameworks, harnessing sparse experimental insights, proficiently prognosticate protein fitness with remarkable accuracy via techniques such as zero-shot and few-shot learning. Zero-shot paradigms, typified by methodologies like VAEs and Transformers, adeptly gauge the likelihood of a novel protein sequence’s functionality by discerning patterns from naturally occurring counterparts.
Conversely, supervised regression frameworks encompassing deep learning and ensemble methodologies leverage labeled data to forecast fitness landscapes and steer the quest for optimal sequences. Active learning strategies further hone this process by striking a balance between exploration and exploitation, leveraging uncertainty quantification models such as Gaussian processes to traverse the fitness landscape more adeptly. This iterative modus operandi, entailing the amalgamation of machine learning prognostications with experimental validation, assumes pivotal significance in realizing optimal solutions in protein engineering endeavors.
Conclusion:
The integration of machine learning in protein engineering heralds a new era of efficiency and innovation across various sectors. By leveraging extensive data and advanced algorithms, businesses can anticipate faster drug discovery, enhanced agricultural practices, and more sustainable solutions, thereby driving growth and competitiveness in the market.